Structure search
You can perform a structure search if you are looking for molecules with a specific pattern or a fragment. This tool is useful when navigating many samples.
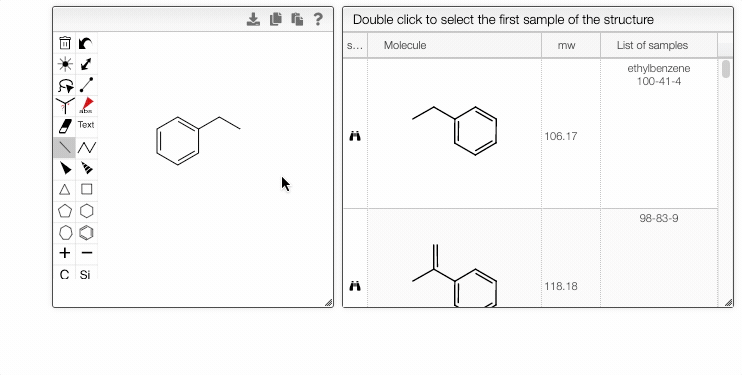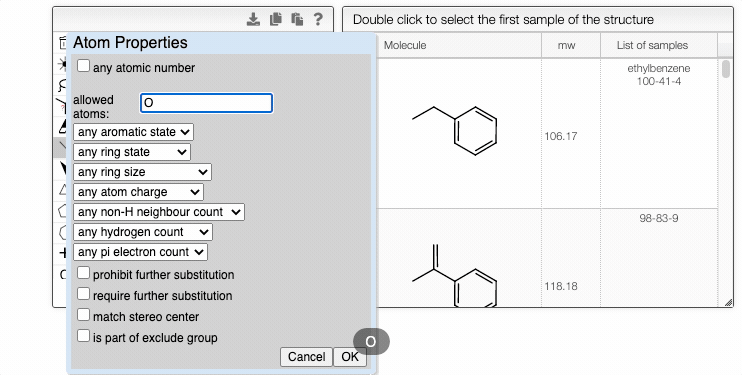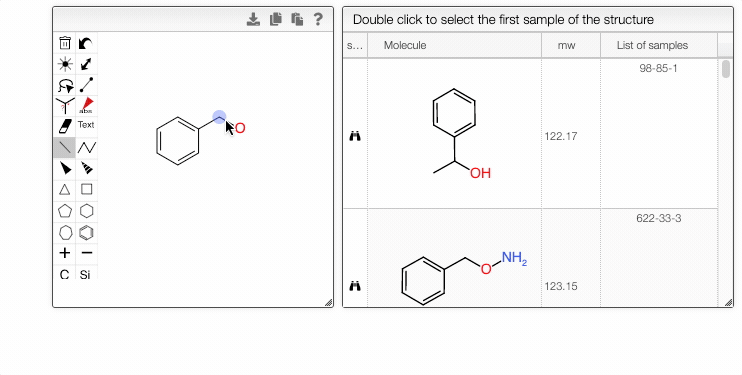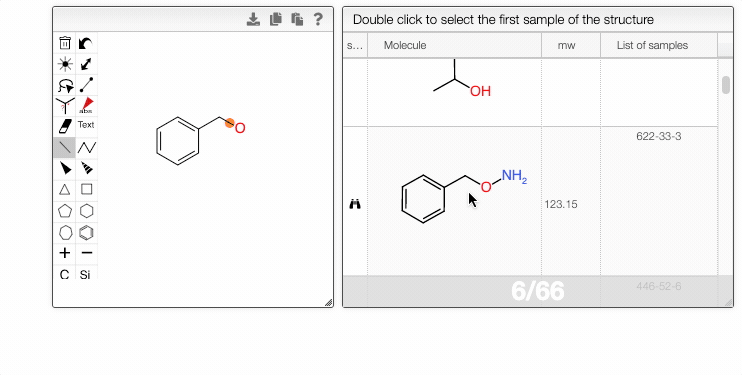
When you select a sample from the Home page and click the Structure search module, the chosen molecule will be searched by default. Alternatively, you may modify the structure in the OCL editor. The matching structures will be available in the tab on the right.
If a molecule appears in more than one sample, you can choose the entry in the List of samples tab.
- The right module displays the list of samples associated with the selected structure. Double-clicking a row will take you back to the main tab, with the corresponding sample selected in the List of selected samples.
- Double-clicking a row in the center module will take you directly to the first matching sample.
There are two available search modes: by substructure and by similarity. Both are based on 512 molecule fragments. Each fragment is described by binary numbers, representing the presence or absence of certain attributes.
In the case of substructure search, the matching molecules contain at least those fragments that were in the query substructure. The structure search uses the same algorithms implemented in Datawarrior. They are open source and available as part of openchemlib (https://github.com/actelion/openchemlib, https://github.com/cheminfo/openchemlib-js).
The similarity search is based on Tanimoto algorithm. It uses the cumulated fragment value and compares the distance between two molecules in the descriptor space. The Tanimoto coefficient can take values between 0, for completely dissimilar molecules, and 1, for identical molecules. Beyond the threshold of 0.85, the molecules are considered similar.
Structure edition is powered by [OCL editor].
Advanced Query Features
Advanced Query features
You can fine-tune the search by specifying atomic and bond properties. These options can be accessed by hovering over the atom or bond of interest and pressing q. For example, you can allow certain atoms at this specific position, or you can modify the ring size.
Furthermore, you can include separate molecules in the search. This will result in structures containing both fragments with no restrictions on orientation or connectivity. One of the molecules can be selected and excluded from the search. It removes the structures completely from the search results. The excluded fragment is highlighted in pink.
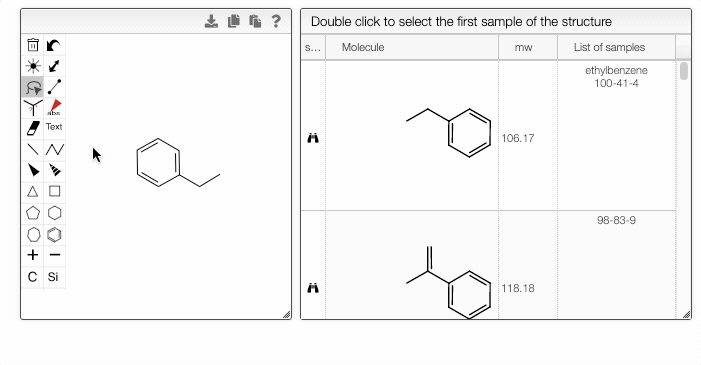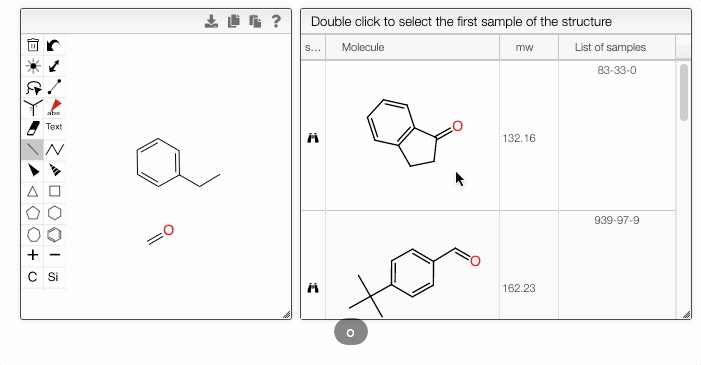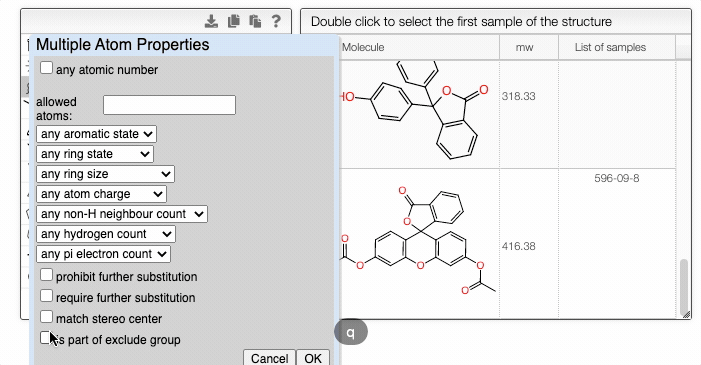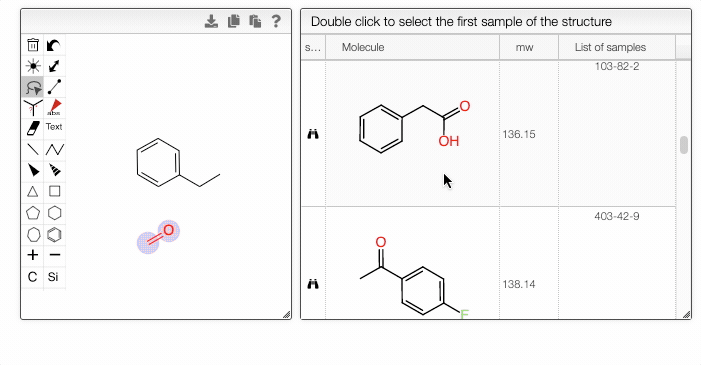
In addition, you can use the dropdown selector to choose whether to search a structure that contains all the fragments, a structure that contains any fragments, or by similarity.

It is also possible to search for structures that contains experimental spectra. The following fields can be used in the search:
nbNmr,nb1d,nb2d,nb13c,nb1h: number of spectrum registered for NMR, 1d NMR...nbMass,nbIR,nbTGA,nbDSC,nbRaman,nbMass,nbUV,nbXray: number of spectrum registered for IR, Raman ...mf,mw,em: molecular formula, molecular weight and exact mass (monoisotpic mass)title: title of the sample.meta: meta information.owner: name of the owner.modified: last modification date of the sample.created: creation date of the sample.
For example, you can draw a substructure and search for all the samples that contain at least one 1D NMR spectrum with the query nbNmr:>0.
Stock status
You can also search for the stock status of structures by using the status field. For example, by querying status:500, you can find all structures with a stock status released. This allows you to filter samples based on their availability or stock level.
Location
You can also search for the location of structures by using the location field. For example, by querying location:CH_123, you can find all structures with a location CH_123. This allows you to filter samples based on their physical location or storage status.