Cyclic voltammetry
Introduction
Cyclic voltammetry is a type of potentiodynamic electrochemical measurement. A cyclic voltammetry graph is a graph of the current as a function of the potential. Both of these variables depend on time.
This tool allows the importation of data as a txt or a JCAMP file.
Upload a file
How to upload a file
In this view, it is possible to upload one or more spectra of a target sample by drag/drop either a JCAMP-DX or a text file. An individual spectrum can be visualized and analyzed by clicking on its name in the "List of files". Several spectra can also be compared. The information about the chosen sample can be found in the colored window on the right.
Upload spectrum
You can upload a spectrum in 3 different ways:
- by dragging your files and then dropping them in the designated zone.
- by clicking on the upload zone and selecting the files to upload.
- by copying the spectrum into the clipboard and then pasting it using CTRL+V (or ⌘+V on MacOS).
The format of the file to be uploaded should be JCAMP-DX (extension .dx or .jdx), this is the usual format used on most IR spectrophotometers.
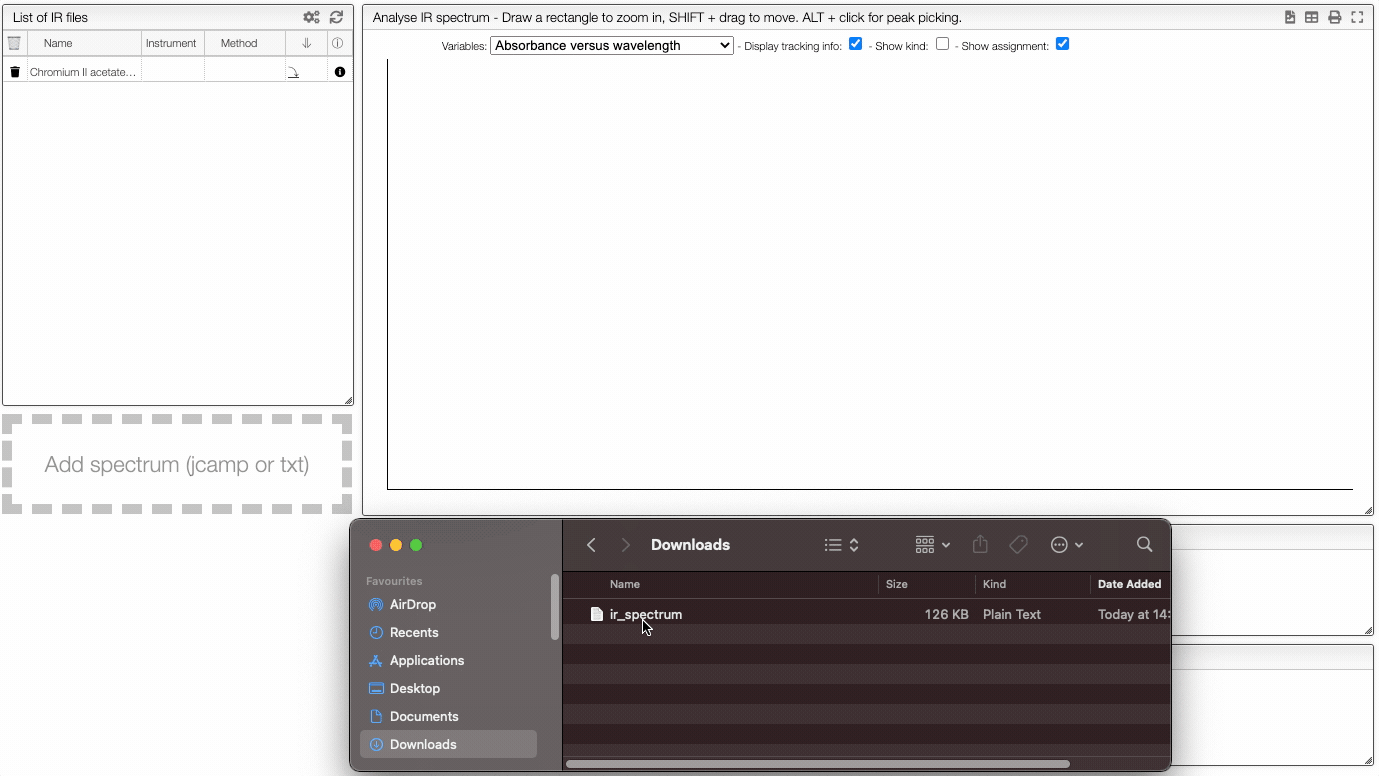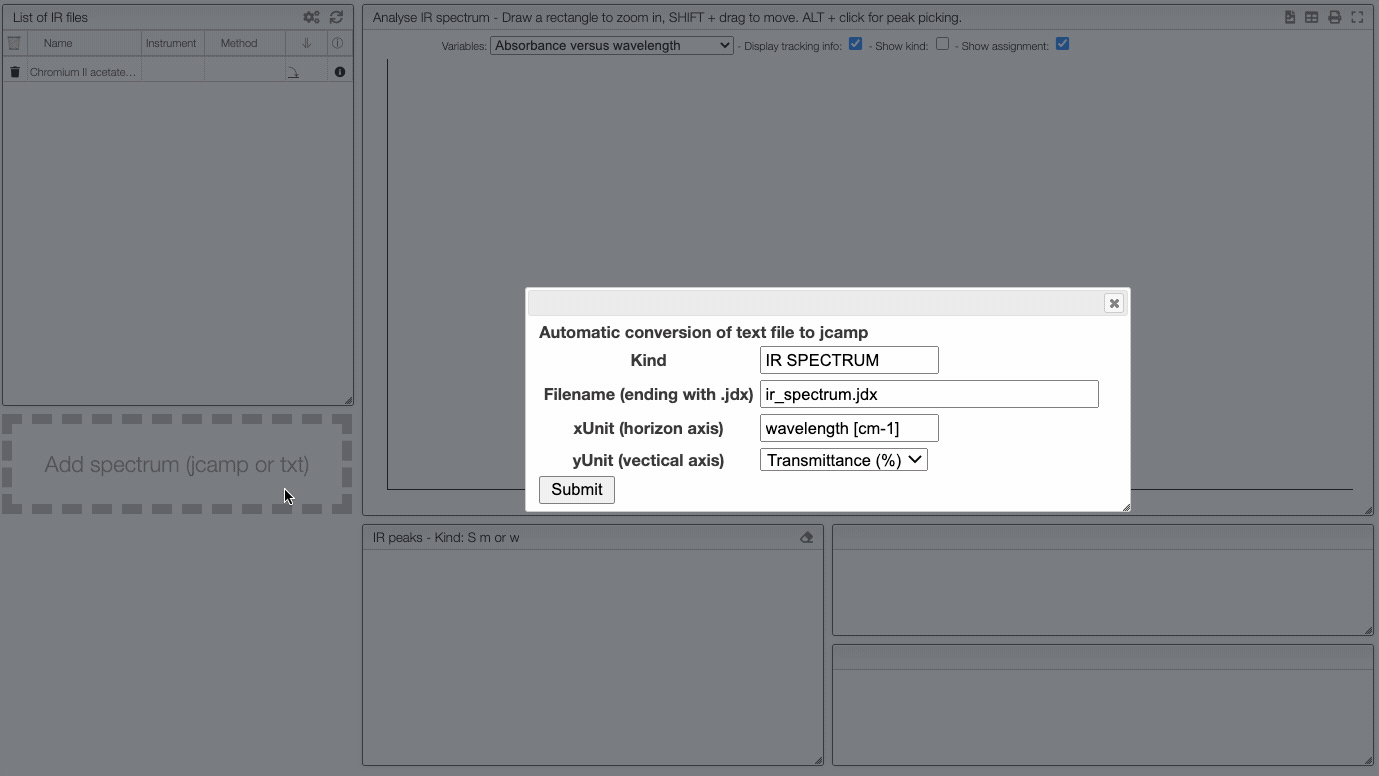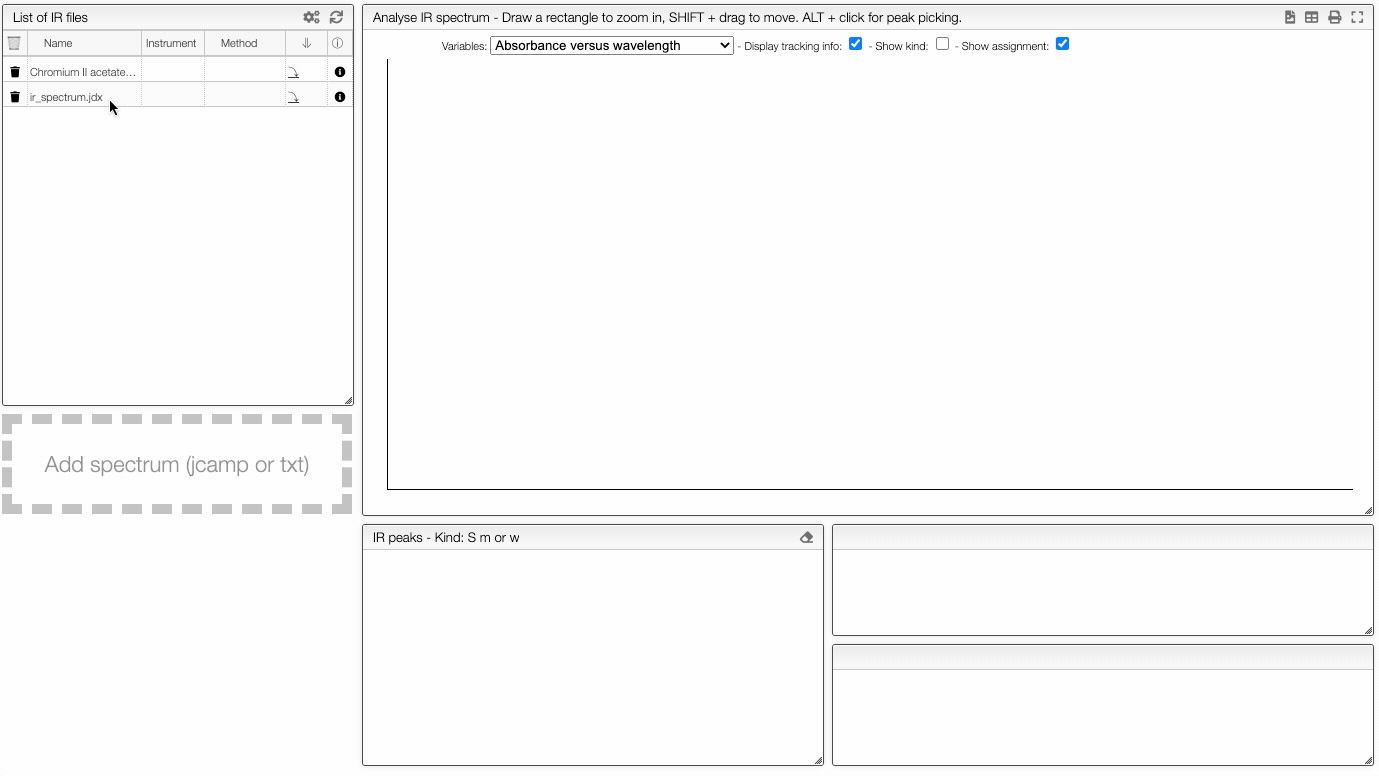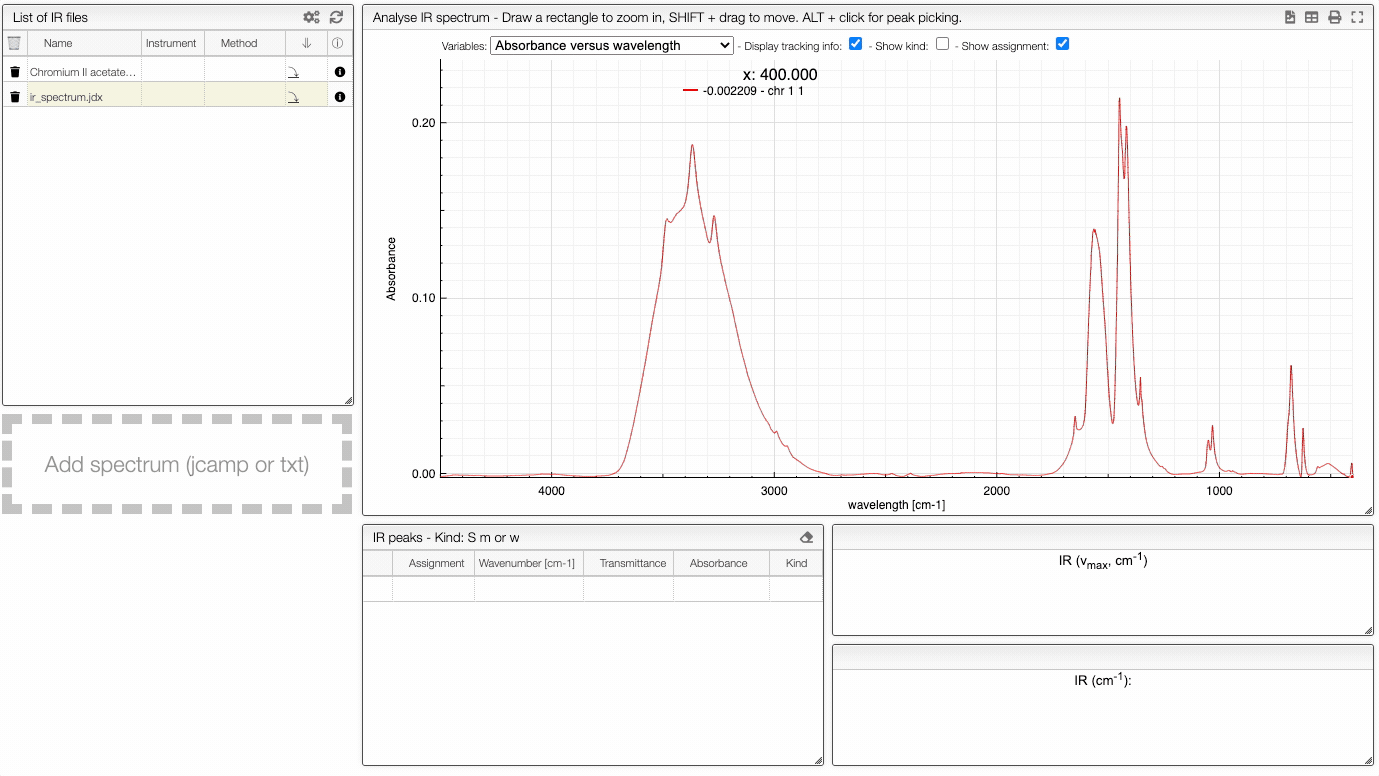
If you click on an imported file, the spectrum is plotted on the middle panel as following. You can manipulate it.
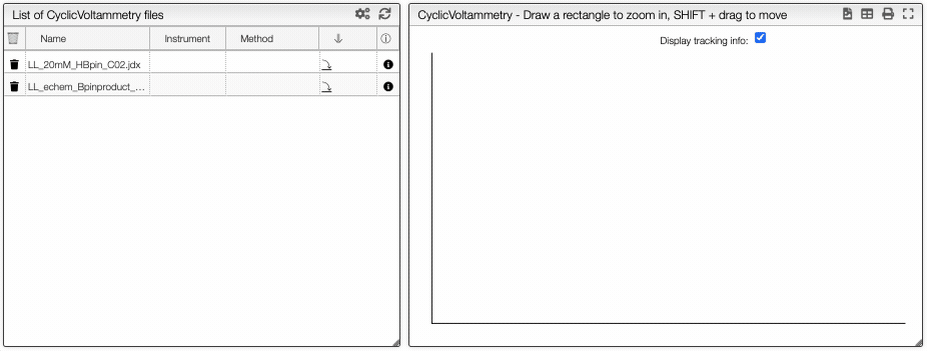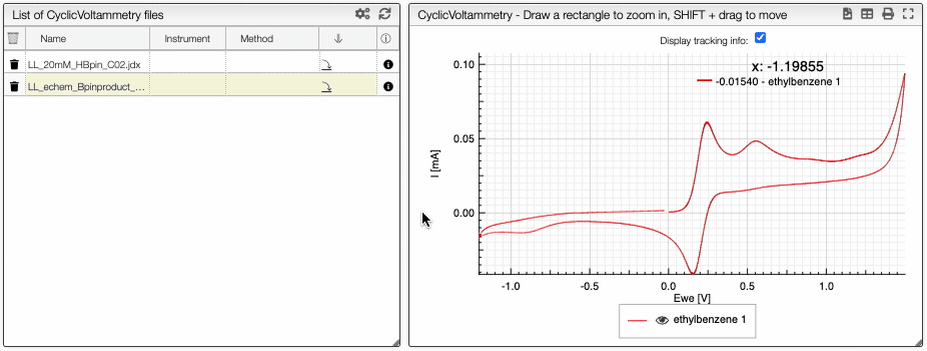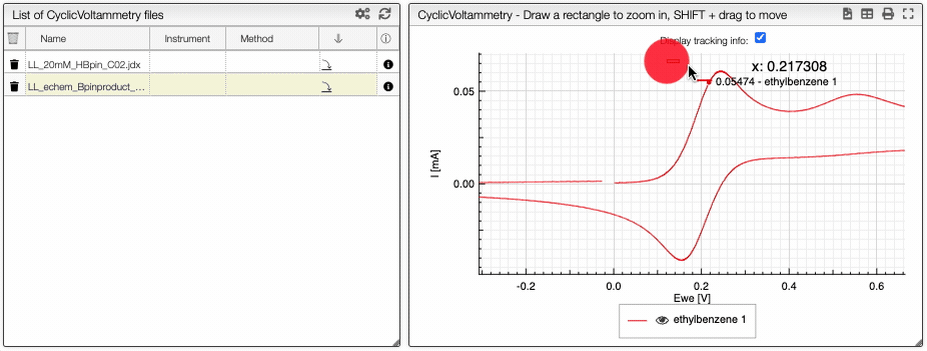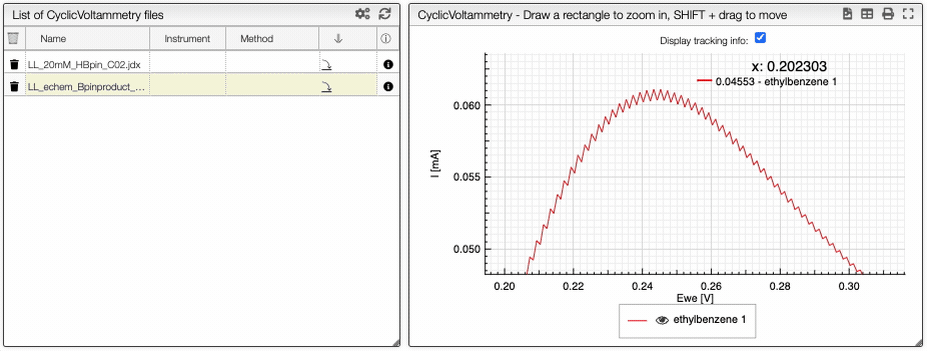
Preprocessing
On the right panel, you can apply mathematical operations to the spectra with the preprocessing module.
Apply mathematical tools to the spectra.
Preprocessing
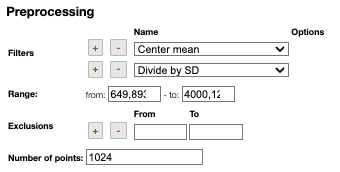
Filters
You can apply the following filters to the spectra to enhance the visualization. The modifications include the following:
Center Mean: subtract the mean from every variable observation in the dataset, so that the new variable's mean is centered at 0.Center Median: subtract the median from every variable observation in the dataset, so that the new variable's median is centered at 0`Divide by SD: divide every variable observable in the dataset by the standard deviation yields a distribution with a standard deviation equal to 1.Normed: Specify a value in thevaluefield and select the type of normalization:Sum to value: normalize the integral under the curve so that it sums to the specified value.Absolute sum to value: normalize the integral under the curve so that the absolute sum sums to the specified value.Max to value: normalize the maximum value to the specified value.
Rescale (x to y): rescale the graph such that the y-values fit between specified minimum and maximum values.First derivative: calculate the first derivative of the spectra.Second derivative: calculate the second derivative of the spectra.Third derivative: calculate the third derivative of the spectra.Savitzky-Golay: smooth the spectra and calculate derivatives based on the following parameters:Window: smoothing window size, must be an odd number, greater than 5.Derivative: derivative order.Polynomial: the degree of the polynomial used to calculate the Savitzky-Golay.
AirPLS baseline: baseline correction using adaptive iterative reweighed penalized least squares algorithm.Iterative polynomial baseline: baseline correction using iterative polynomial fitting algorithm.Rolling average baseline: baseline correction using a rolling average.Rolling median baseline: baseline correction using a rolling median.Rolling ball baseline: baseline correction using a rolling ball.Ensure growing X values: ensure that the x-values are in increasing order.Function on X: apply a function to the x-values. For example,log(x).Function on Y: apply a function to the y-values. For example,log10(y+1).Calibrate X: calibrate the x-values with the parametersfrom,to,nbPeakandtargetX.Pareto normalization: Pareto scaling, which uses the square root of standard deviation as the scaling factor, circumvents the amplification of noise by retaining a small portion of magnitude information. 10.1016/j.molstruc.2007.12.026
One classical preprocessing algorithm is Standard Normal Variate (SNV). This preprocessing can be achieved by selecting the 2 options Center mean and Divide by SD.
Selecting the range
A certain range of x-values can be selected to show only a part of the spectrum using Range.
Exclusions
Depending on the analysis, some regions should be removed using Exclusions in order to improve the visualization.
Number of points
Number of points can be changed to reduce the number of points in the spectra.
Compare spectra
You can compare multiple spectrum by clickling on the Compare button. It is then possible to hide/show spectra.
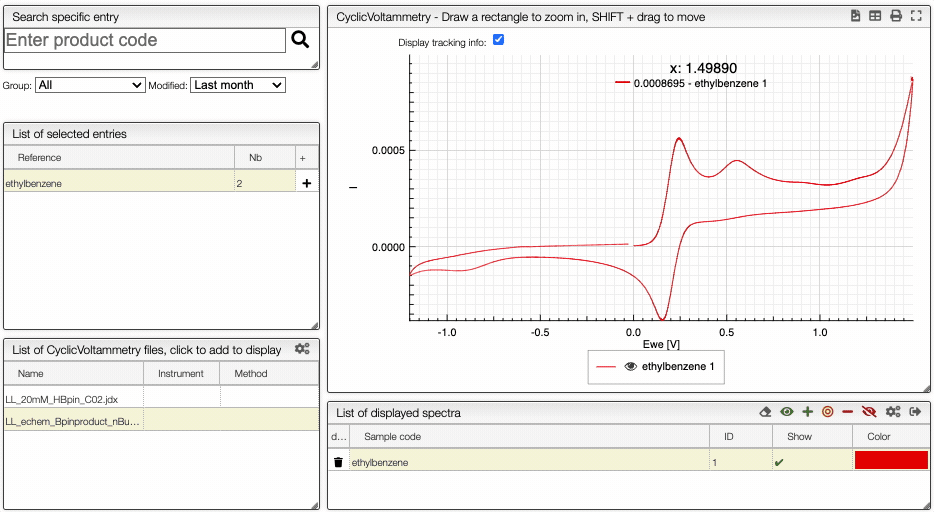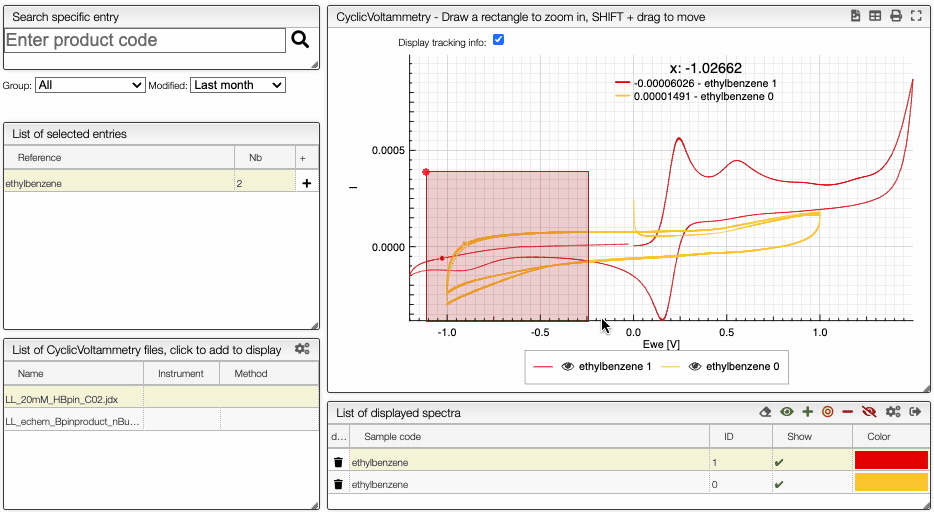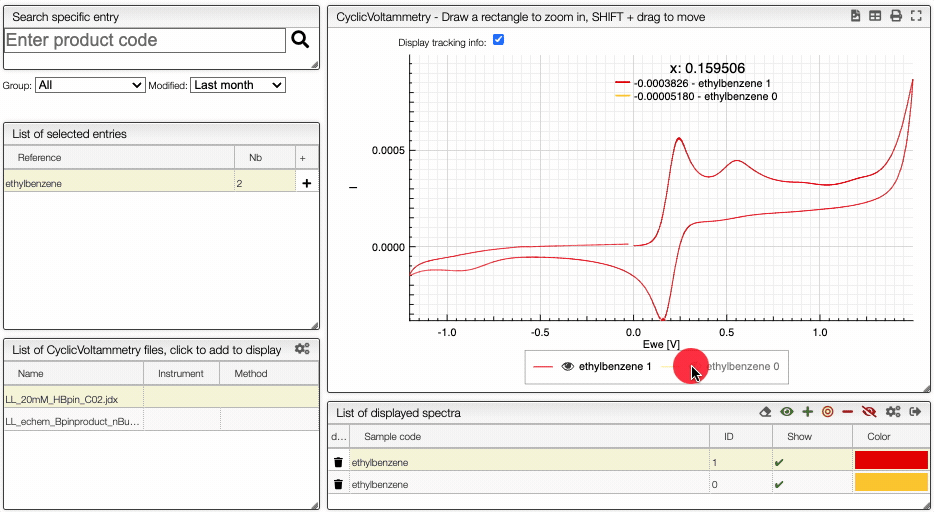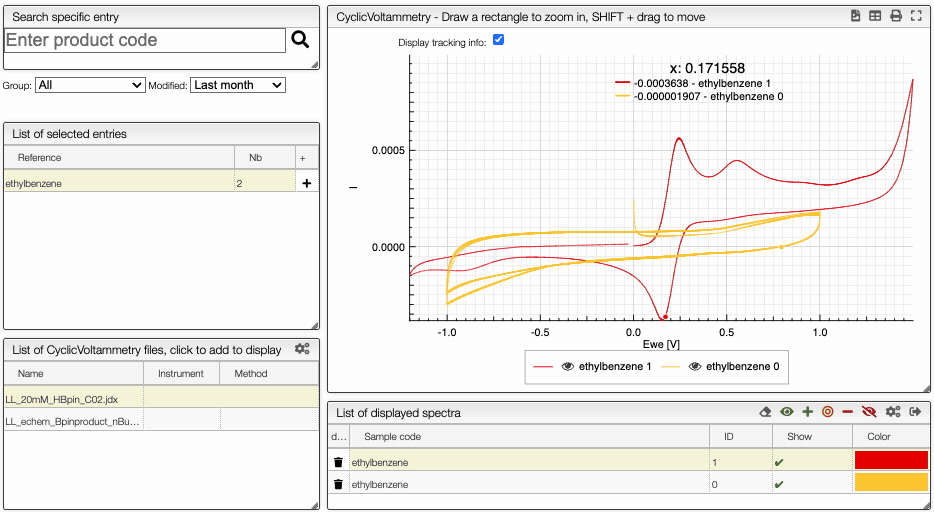
Learn more about cyclic voltametry: 10.1021/acs.jchemed.7b00361
In many tables it is possible to select which columns to display. This is achieved by clicking on the icon.
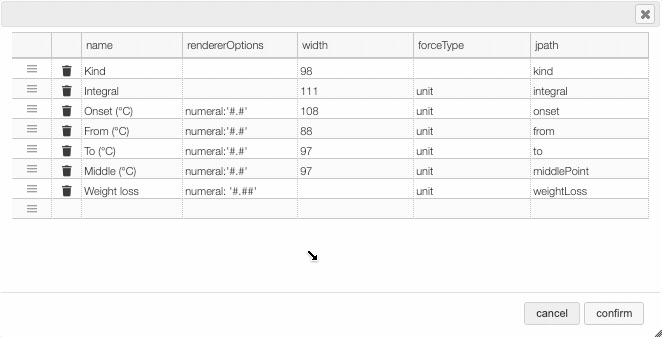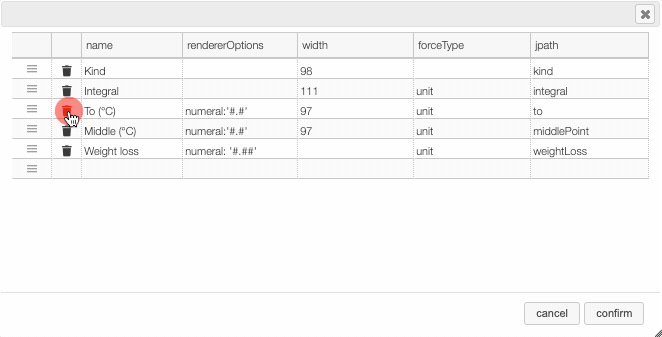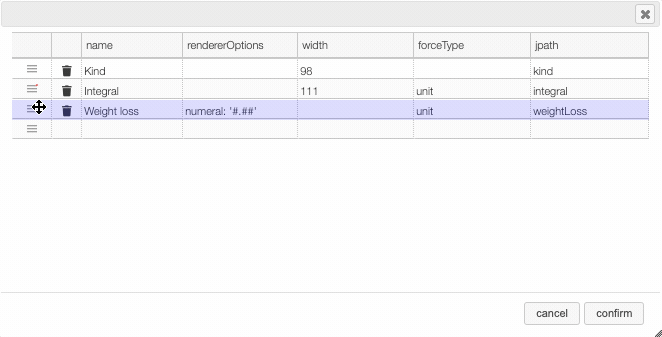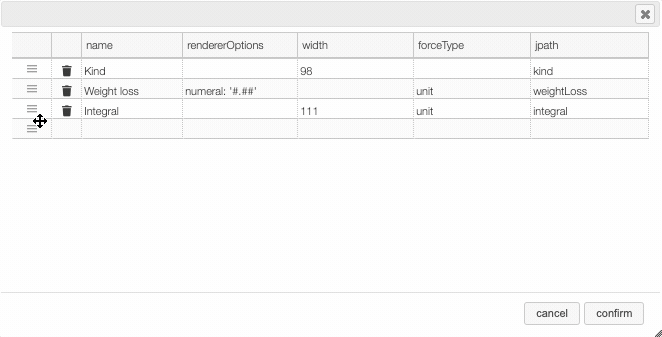
After clicking on the icon a dialog box opens that allows to add a new column.
There are 5 parameters to fill for a new column:
- name: the column name. This will be displayed as the header to the column.
- rendererOptions: options that allows among other to format numbers. One very useful formatter is
numeral(see after) - width: number of pixels for the specific column. May stay empty for automatic layout.
- forceType: select how to display complex values (see later)
- jpath: where to find the information to display in the column
In order to add a new column you need to select the jpath using the hierarchical drop down menu.
Columns can be moved or rearranged as well.
rendererOptions: numeral
number,
| Value to format | rendererOptions | Result |
|---|---|---|
| 12.345678 | numeral:'#.##' | 12.34 |
| 12.3 | numeral:'#.00' | 12.30 |
| 0.3 | numeral:'#.0 %' | 30.0 % |
forceType
In the database some values are stored as an object that needs to be displayed to the user in an intuitive way.
For example the unit type will store in the database the value using as units SI (we convert the data to the units defined by the 'standard international') and specify in which units the user would like to display the data. So in the following example we are in fact storing the value -150°C by storing the value in SI (Kelvin) and specifying that the user wants to see it in °C.
{
"SI": 123.15,
"unit": "°C"
}
Another example is the valueUnits type that will store the data in 2 different properties
(value and unit). In this case the value is stored in the specified units and there is no conversions.
{
"value": 123,
"units": "°C"
}
While this way to store the data in the database is very practical it is not the way that the user would like to see his results. We have therefore the possibility to forceType: define how the user would like to see the results.
Other types include mf. This formatter allows to correctly display a molecular formula that is stored in the database as "C10H20O3". i.e. it will put the numbers in subscript (C₁₀H₂₀O₃).