13C prediction
This tool allows you to predict the 13C NMR spectrum of your sample or any other molecule.
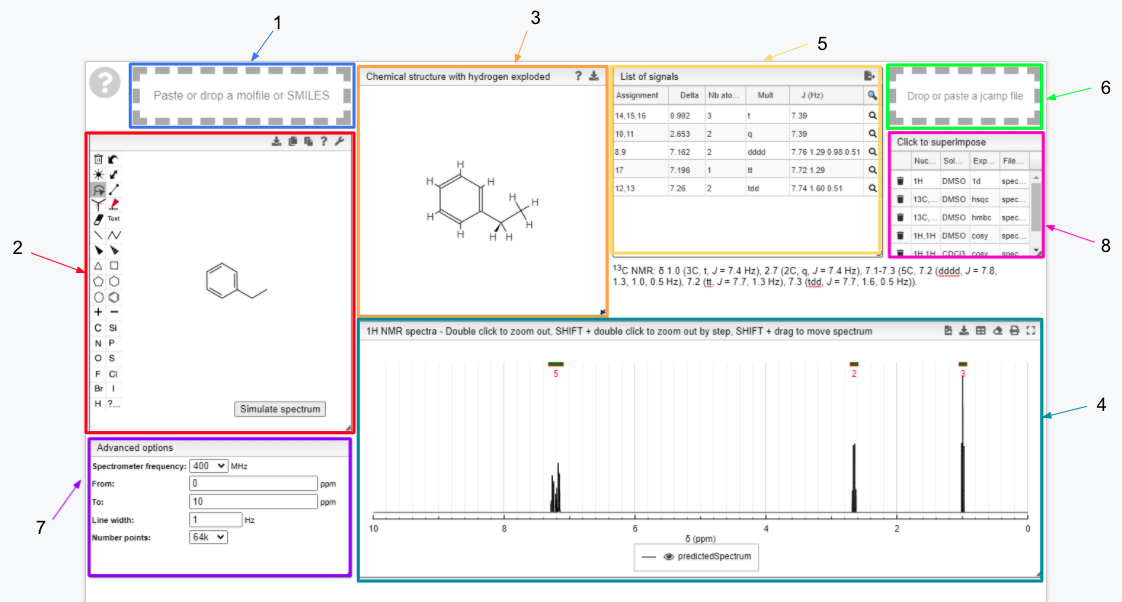
- Drag and drop module - paste a molfile or a SMILES string of a molecule
- Draw a chemical structure and predict module - draw the structure of the molecule
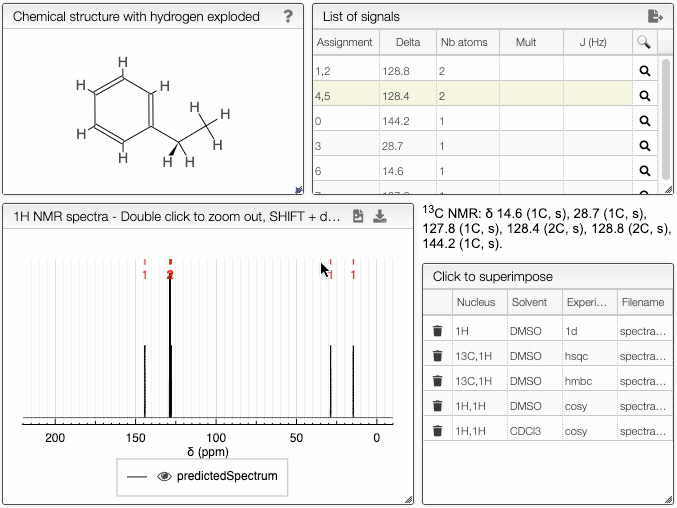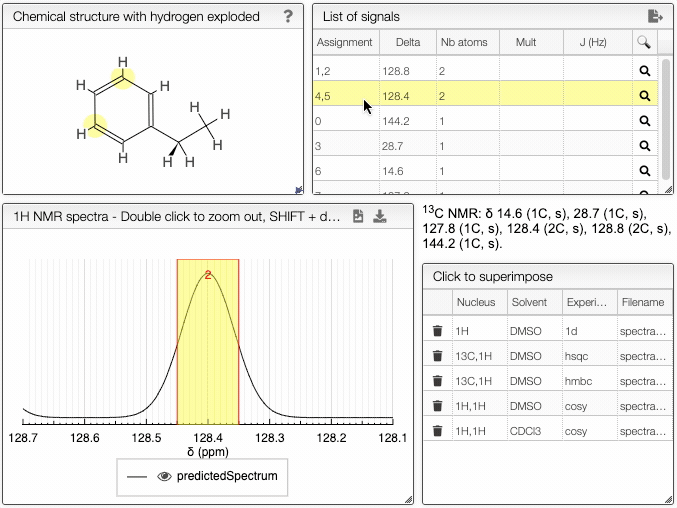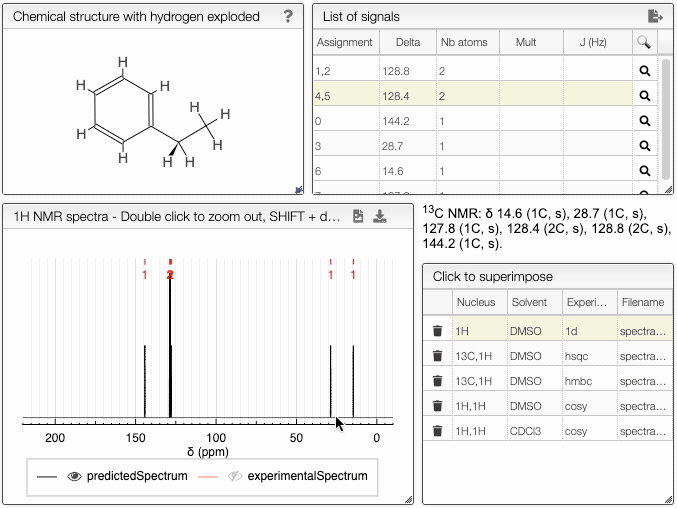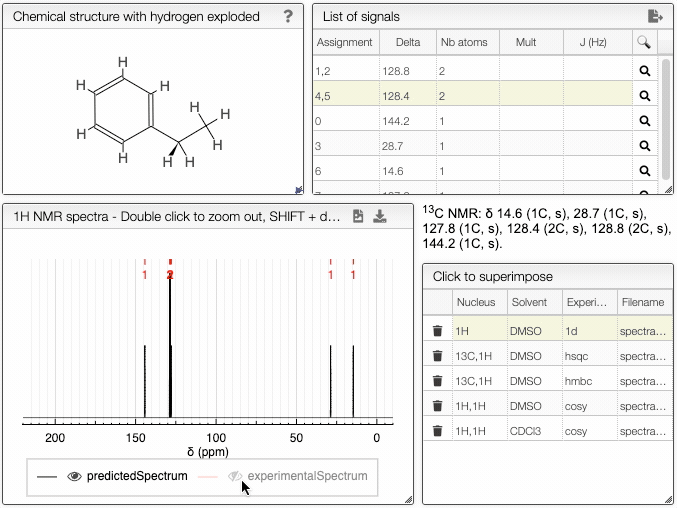
- Chemical structure with explicit hydrogens module - explicit representation of hydrogens in a molecule
- 13C NMR spectrum module - predicted 13C spectrum of the chosen molecule
- Signal module - list of obtained peaks and the corresponding chemical shifts
- Drag and drop module - paste a JCAMP-DX of an experimental spectrum for comparison
The structure of the currently selected sample will be already drawn so that you may go ahead and simulate its spectrum right away. If you wish to simulate the spectrum of another molecule, you can draw it or you can paste the structure in the form of a molfile or a SMILES string. Structure drawing is powered by JSME. You may also drop or paste a JCAMP-DX file to superimpose an experimental spectrum over the prediction.
How is a JCAMP-DX file structured.
JCAMP-DX file format
JCAMP-DX (Joint Committee on Atomic and Molecular Physical Data Exchange) is a standard file format for the exchange of spectra and related physical and chemical information between different spectrometers, databases or other systems.
The information is stored using ASCII characters and the file can be viewed, corrected and annotated with a text editor. The spectra are stored as a table containing (x,y) coordinate pairs. Besides the data points, it is possible to store metainformation and make comments. The file extension is .jdx.
A JCAMP-DX document is composed of an unlimited number of Labelled Data Records (LDRs). Each LDR starts with a “##” and ends with “=”. Any space, comma, slash or hyphen is removed and the text is written with capital letters.
Some examples of Data Labels:
- TITLE : title of the experiment
- END : the last line of the file
- XUNITS : the units reported on the x-axis
- NPOINTS : number of points
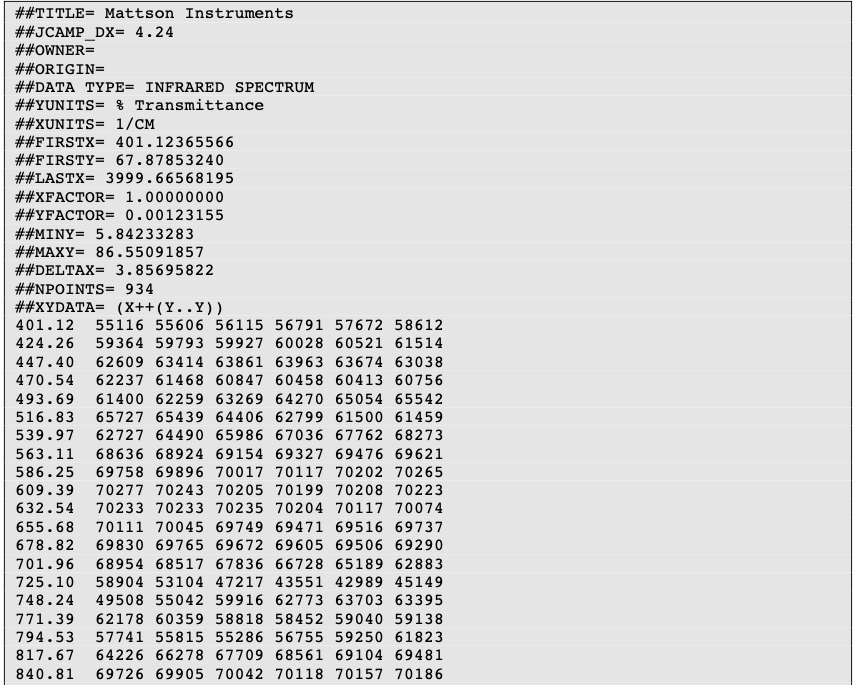
Two important LDRs are “XYDATA” and “PEAKTABLE”, which contain the spectral information. The former gives information in the form of a table where the first value in a line stands for an x coordinate and any subsequent values are y-coordinates with an equidistant increment on the x-axis. The latter provides information as a collection of (X,Y) pairs.
It is commonplace to compress the data tables. For instance, the table of numbers can be replaced by a line of characters (pseudo-digits). Among these pseudo-digits, there are PAC, SQZ, DIF, DIFDUP.

An example of compressed data using DIFDUP

An in depth description is given in the original paper by McDonald and Wilks. Insofar as JCAMP-DX is a well-described and accessible format, it partially aligns with the FAIR (Findable, Accessible, Interoperable, Reusable) principles . It is interoperable and reusable. Provided that the user makes it findable and accessible, JCAMP-DX will fully comply with the aforementioned principles.
The simulated spectrum, the chemical structure with explicit hydrogens and the list of peaks modules are linked, so that hovering over an entry in the list will highlight the corresponding atom in the structure and the relevant peak in the spectrum.
NMR prediction is done with NMRshiftDB. It is an NMR database for organic structures and their spectra. Using this information, in conjunction with the principle of HOSE code, a machine learning model makes chemical shift predictions.
What is the HOSE code.
HOSE code
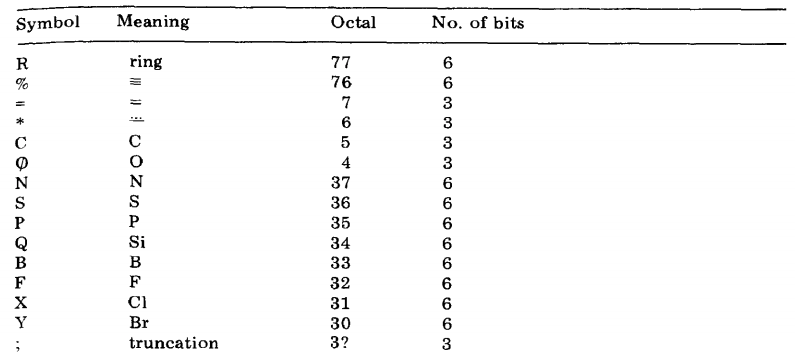
The HOSE (Hierarchically Ordered Spherical Environment) code describes the environment of individual atoms spherically. The environment of a particular atom is described in the form of a string of characters. The priority rules and necessary syntax have been outlined in the original paper in 1978. For instance, here are some symbols.
The prediction of the signal due to each individual atom is done by considering the chemical environment of the atom by layers like onion peels. See the following example.
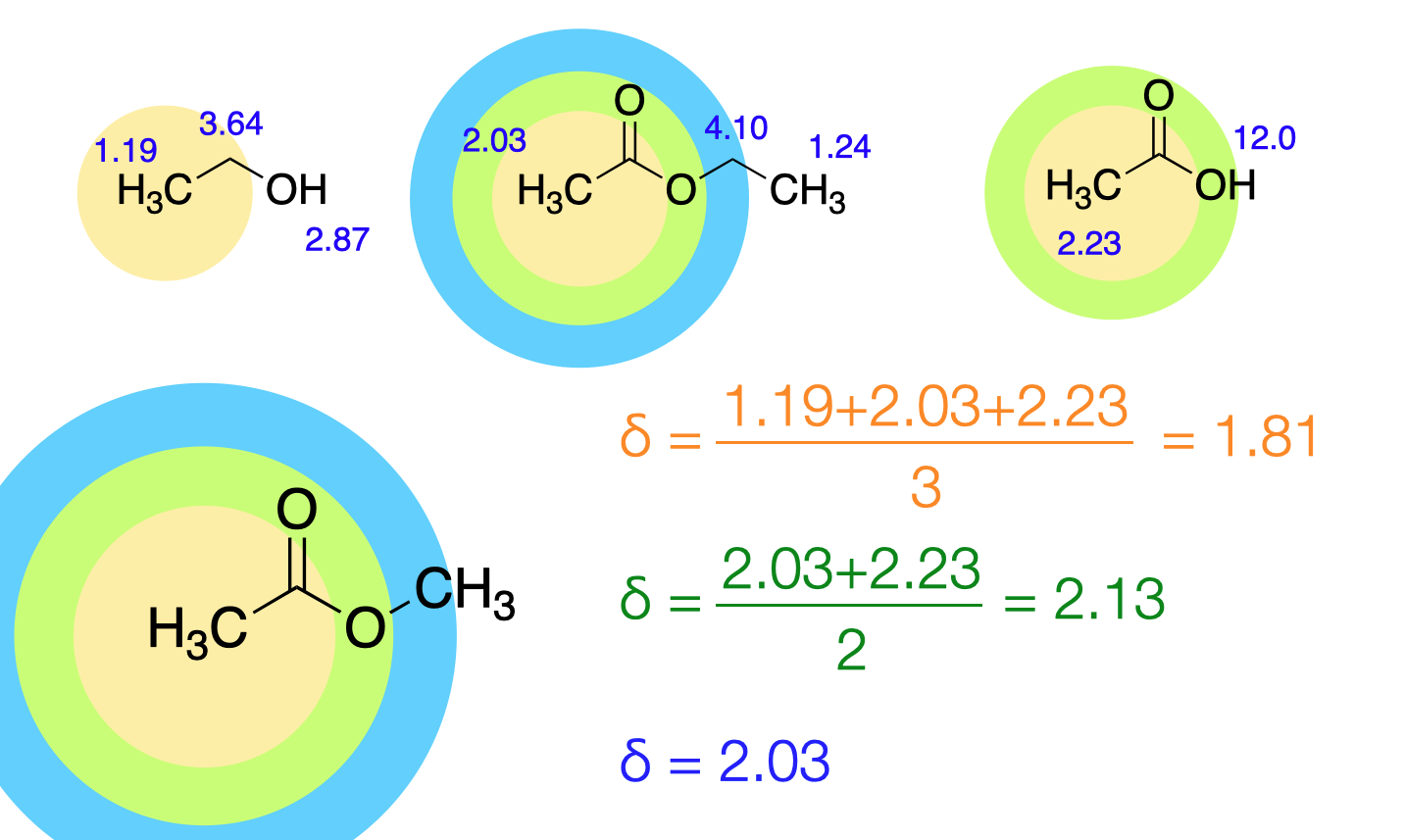
The HOSE code is based on nmrshiftdb.